Lucy Alumna Anna Lappala (Physics 2011) on her latest research combining lab data with supercomputing power to understand chromosome dynamics
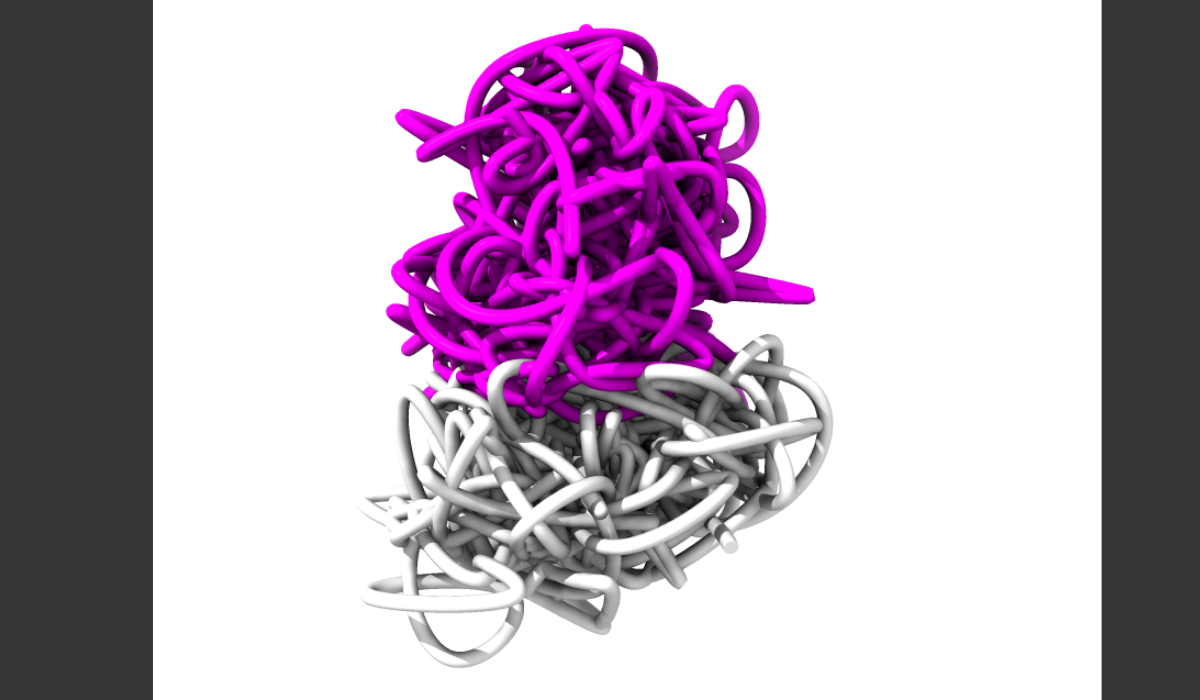
Image: RNA particles swarm an X chromosome from a mouse in a new visualization of X chromosome inactivation. Credit: Los Alamos National Laboratory
Using supercomputer-driven dynamic modeling based on experimental data, researchers can now probe the process that turns off one X chromosome in female mammal embryos. This new capability is helping biologists understand the role of RNA and the chromosome’s structure in the X inactivation process, leading to a deeper understanding of gene expression and opening new pathways to drug treatments for gene-based disorders and diseases.
Lucy alumna Dr Anna Lappala, a polymer physicist at Massachusetts General Hospital and the Harvard Department of Molecular Biology and a visiting scientist at Los Alamos National Laboratory, is first author of the paper published on 4th October in the Proceedings of the National Academy of Sciences (PNAS).
The paper, titled 4D chromosome reconstruction elucidates the spatial reorganization of the mammalian X-chromosome, is available at doi.org/10.1073/pnas.2107092118
“We used supercomputers to model how the mammalian X chromosome folds and how its genes are being deactivated in the process. The model is based on experimental data and reveals a number of interesting features, such as the origami-like folding pattern of chromosomes and their glass-like behavior” comments Dr Lappala.
“This is the first time we’ve been able to model all the RNA spreading around the chromosome and shutting it down. From experimental data alone, which is 2D and static, you don’t have the resolution to see a whole chromosome at this level of detail. With this modeling, we can see the processes regulating gene expression, and the modeling is grounded in 2D experimental data from our collaborators at Massachusetts General Hospital and Harvard.”
The model, called 4DHiC - considered 4D because it shows motion, including time as the fourth dimension - runs on Los Alamos supercomputers. It incorporates experimental data from the genome obtained through a molecular method called Chromosome Conformation Capture (HiC). The combined molecular and computational methodology allowed researchers to look into a complex dynamical process of X chromosome inactivation, showing how the chromosome reorganizes itself in 3D, effectively turning off gene expression.
Based on the work in this paper, Los Alamos is currently developing a Google Earth-style browser where any scientist can upload their genomic data and view it dynamically in 3D at various magnifications, said Karissa Sanbonmatsu, a structural biologist at Los Alamos National Laboratory, corresponding author of the paper, and a project leader in developing the computational method.
About PNAS
PNAS is one of the world's most-cited and comprehensive multidisciplinary scientific journals, publishing more than 3,500 research papers annually.
The Proceedings of the National Academy of Sciences (PNAS), the official journal of the National Academy of Sciences (NAS), is an authoritative source of high-impact, original research that broadly spans the biological, physical, and social sciences. The journal is global in scope and submission is open to all researchers worldwide.
About Dr Anna Lappala
Anna worked on her PhD in Physics under Prof. Eugene Terentjev of Cavendish Laboratory. During her time at Lucy, she was the captain of the Cambridge University Kickboxing Society, received a Half Blue in the sport (she successfully proposed a University - wide regulation that allowed women to receive Half Blues in kickboxing), and loved playing the piano at Lucy. After graduating with a PhD, she was a Center for Nonlinear Studies Fellow at Los Alamos National Laboratory in New Mexico, and she is currently an Instructor at Harvard.